STEPHANOPOULOS LAB
SELF-ASSEMBLING
BIONANOTECHNOLOGY

PUBLICATIONS
As an ASU faculty member:
(*= corresponding author)
Site-specific modification of synthetic peptides with two orthogonal DNA handles. A. Novacek, B. Ugaz, N. Stephanopoulos*. (manuscript in preparation)
High-affinity binding to the SARS-CoV-2 spike trimer by a multivalent protein-DNA synthetic antibody. Y. Xuǂ, R. Zhengǂ, A. Prasadǂ, M. Liu, X. Zhou, R. Porter, M. Sample, E. Poppleton, J. Procyk, H. Liu, H. Lee, H. Yan*, P. Sulc*, N. Stephanopoulos*. (submitted; pre-print available on bioRxiv: ǂ co-first authors)
Structure-based discovery of hydrocarbon-stapled paxillin peptides that block FAK scaffolding in cancer. L. Reyes, L. Naser, W.S. Weiner, D. Thifault, E. Stahl, L. McCreary, R. Nott, C. Quick, A. Buchberger, C. Alvarado, A. Rivera, J.A. Miller, R. Khatiwala, B.R. Cherry, R. Nelson, J.M. Garcia, N. Stephanopoulos, R. Fromme, P. Fromme, W. Cance, T. Marlowe*. Nat. Commun. (2025). 16:2060.

Geometrically constrained cytoskeletal reorganization modulates DNA nanostructures uptake. P. Elblova, H. Andelova, M. Lunova, J. Anthi, S.J.W. Henry, X. Tu, A. Dejneka, M. Jirsa, Nicholas Stephanopoulos*, and Oleg Lunov*. J. Mater. Chem. B. (2025) 13:2335-2351.

Click chemistry-enabled functionalization of cellulose nanocrystals with single-stranded DNA for directed assembly. D. Bukharina, K. Cauffiel, L.M. Killingsworth, V. Poliukhova, M. Kim, J. Brower, J. Bernal-Chanchavac, N. Stephanopoulos, V.V. Tsukruk*. ACS Biomat. Sci. Eng. (2024) 10:6155–6166.

Peptide-coated DNA Nanostructures as a Platform for Control of Lysosomal Function in Cells. P. Elblova, M. Lunova, S.J.W. Henry, X. Tu, A. Dejneka, J. Havelkova, Y. Petrenko, M. Jirsa, Nicholas Stephanopoulos*, and Oleg Lunov*. Chem. Eng. J. (2024) 485:155633.

Templating peptide chemistry with nucleic acids: towards artificial ribosomes, cell-specific therapeutics, and novel protein-mimetic architectures. A. Novacek, B. Ugaz, N. Stephanopoulos*. Biomacromolecules (2024) 25:3865–3876.

Inverse design of a pyrochlore lattice of DNA origami through model-driven experiments. H. Liu, M. Matthies, J. Russo, L. Rovigatti, R.P. Narayanan, T. Diep, D. McKeen, O. Gang, N. Stephanopoulos, F. Sciortino, H. Yan, F. Romano, P. Sulc*. Science (2024) 384:776.

Construction of reconfigurable and polymorphic DNA origami assemblies with coiled-coil patches and patterns. T. Teng, J. Bernal-Chanchavac, N. Stephanopoulos, C. Castro*. Small (2024) 11:2307257.

DNA-Nanostructure Guided Assembly of Proteins into Programmable Shapes. Q. Lu, Y. Xu, E. Poppleton, P. Sulc, N. Stephanopoulos*, Y. Ke*. Nano Lett. (2024) 24:1703.

Using dynamic biomaterials to study the temporal role of bioactive peptides during osteogenesis. F.M. Fumasi, T. MacCulloch, J. Bernal-Chanchavac, N. Stephanopoulos, J.L. Holloway*. Biomater. Adv. (2024) 157:213726.

CytoDirect: a DNA nanodevice for specific and efficient delivery of functional payloads to the cytoplasm. L. Yu, Y. Xu, M. Al-Amin, S. Jiang, M. Sample, A. Prasad, N. Stephanopoulos, P. Sulc, H. Yan*. J. Am. Chem. Soc. (2023) 145:27336.


High-affinity host–guest recognition for efficient assembly and enzymatic responsiveness of DNA nanostructures. R.P. Narayanan, A. Prasad, A. Buchberger, L. Zou, J. Bernal-Chanchavac, T. MacCulloch, N.E. Fahmi, H. Yan, F. Zhang M.J. Webber*, N. Stephanopoulos*. Small (2023). 20:2307585.

Site-specific arrangement and structure determination of minor groove binding molecules in self-assembled three-dimensional DNA crystals. C.R. Simmons, A. Buchberger, S.J.W. Henry, A. Novacek, N.E. Fahmi, T. MacCulloch, N. Stephanopoulos*, H. Yan*. Am. Chem. Soc. (2023) 145:26075.

Calcium-triggered DNA-mediated membrane fusion in synthetic cells. Y.-Y. Hsu, S.J. Chen, J. Bernal-Chanchavac, B. Sharma, H. Moghimianavval, N. Stephanopoulos, and A.P. Liu*. Chem. Commun. (2023), 59:8806.

Self-assembly of hybrid peptide-DNA nanostructures using homotrimeric coiled-coil/nucleic acid building blocks. A. Buchberger, M Al-Amin, C.R. Simmons, N. Stephanopoulos*. ChemBioChem (2023), 17:e202300223. (Featured cover article)
Atomistic picture of opening−closing dynamics of DNA Holliday junction obtained by molecular simulations. Z. Zhang, J. Sponer, G. Bussi, V. Mlynsky, P. Sulc, C.R. Simmons, N. Stephanopoulos, M. Krepl*. J. Chem. Inf. Model. (2023), 63, 2794-2809.
Self-Assembling Biomaterials from Proteins, Peptides, and DNA. N. Stephanopoulos*, R. Freeman, H. Yan. ACS Applied Bio Materials (2022), 5:4579. (Guest Editor editorial for special issue)

Bioactive Fibronectin-III10-DNA Origami Nanofibers Promote Cell Adhesion and Spreading. A. Buchbergerǂ, K. Rikerǂ, J. Bernal-Chanchavac, R.P. Narayanan, C.R. Simmons, N.E. Fahmi, R. Freeman*, N. Stephanopoulos*. ACS Appl. Bio. Mater. (2022) 5:4625.

Coarse-grained simulations for the characterization and optimization of hybrid protein-DNA nanostructures. R.P. Narayananǂ, J. Procykǂ, P. Nandi§, A. Prasad§, Y. Xu§, E. Poppleton, D. Williams, F. Zhang, H. Yan, P.-L. Chiu*, N. Stephanopoulos*, P. Sulc*. ACS Nano (2022) 16:14086. (ǂ co-first authors; § co-second authors)

Uncovering temporospatial sensitive TBI targeting strategies via in vivo phage display. B.I. Martinez, G. Mousa, K. Fleck, T. MacCulloch, C.W. Diehnelt, N. Stephanopoulos, S.E. Stabenfeldt*. Sci. Adv. (2022) 8:eabo5047. (see Biodesign highlight)
The interactions between DNA nanostructures and cells: A Roadmap for Successful Applications in Biomedicine A critical overview from a cell biology perspective. A. Frtús, B. Smolková, M. Uzhytchak, M. Lunova, M. Jirsa, S.J.W. Henry, A. Dejneka, N. Stephanopoulos*, O. Lunov*. Acta Biomaterial. (2022) 146:10-22.
The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly. C.R. Simmonsǂ, T. MacCullochǂ, M. Krepl, M. Matthies, A. Buchberger, I. Crawford, J. Sponer, P. Sulc, Y. Liu, N. Stephanopoulos*, H. Yan*. Nat. Commun. (2022) 13:3112. (ǂ co-first authors)

Nanoscale structures and materials from the self-assembly of polypeptides and DNA. J. Bernal-Chanchavacǂ, M. Al-Aminǂ, N. Stephanopoulos*. Curr. Top. Med. Chem. (2022) 22:699. (ǂ co-first authors)
The living interface between synthetic biology and biomaterial design. A.P. Liu*, E. Appel, P. Ashby, B. Baker, E. Franco, L. Guo, K. Haynes, N. Joshi, A. Kloxin, P. Kouwer, J. Mittal, L. Morsut, V. Noireaux, S. Parekh, R. Schulman, S. Tang, M. Valentine, S. Vega, W. Weber, N. Stephanopoulos*, O. Chaudhuri*. Nat. Mater. (2022) 21:390-397. (see accompanying editorial)

Proximity-enhanced synthesis of DNA-peptide-DNA triblock molecules. T. MacCulloch, A. Novacek, N. Stephanopoulos*. Chem. Commun. (2022) 58:4044-4047.

Programmable, self-assembled DNA nanodevices for cellular programming and tissue engineering. A. Gangrade*, N. Stephanopoulos, D. Bhatia*. Nanoscale (2021) 13:16834-16846.

Effect of the protein corona on endosomal escape of functionalized DNA nanostructures. B. Smolková, T. MacCulloch, T. Rockwood, M. Liu, S.J.W. Henry, A. Frtús, M. Uzhytchak, M. Lunova, M. Hof, P. Jurkiewicz, A. Dejneka, N. Stephanopoulos*, O. Lunov*. ACS Appl. Mater. Interfaces (2021) 13:46375–46390.
Highly permeable DNA supramolecular hydrogel promotes neurogenesis and functional recovery after completely transected spinal cord injury. T. Yuan, Y. Shao, X. Zhou, Q. Liu, Z. Zhu, B. Zhou, Y. Dong, N. Stephanopoulos, S. Gui*, H. Yan*, D. Liu*. Adv. Mater. (2021) 33:2102428.

Functionalizing DNA nanostructures for therapeutic applications. S.J.W. Henry, N Stephanopoulos*. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. (2021) 13:e1729.

Reversible control of gelatin hydrogel stiffness using DNA crosslinkers. A. Buchbergerǂ, H. Sainiǂ, K.R. Eliatoǂ, R. Merkley, Y. Xu, A. Zare, J. Bernal, R. Ros*, M. Nikkhah*, N. Stephanopoulos*. ChemBioChem (2021) 22:1755–1760. (ǂco-first authors; selected as a “Very Important Paper” by the journal)

DNA nanodevices as mechanical probes of protein structure and function. N. Stephanopoulos*, P. Sulc. Appl. Sci. (2021) 11:2802.

A Self‐Assembled Rhombohedral DNA Crystal Scaffold with Tunable Cavity Sizes and High‐Resolution Structural Detail. C.R. Simmonsǂ, T. MacCullochǂ, F. Zhang, Y. Liu, N. Stephanopoulos*, H. Yan*. Angew. Chem. Int. Ed. (2020) 59:18619-18626. (ǂco-first authors)

Hybrid nanostructures from the self-assembly of proteins and DNA. N. Stephanopoulos*. Chem (2020) 6:364–405.
Reversible Control of Biomaterial Properties for Dynamically Tuning Cell Behavior. F.M. Fumasi, N. Stephanopoulos, J.L. Holloway*. J. Appl. Polym. Sci. (2020) 137:e49058.

Hierarchical Assembly of Nucleic Acid/Coiled-Coil Peptide Nanostructures. A. Buchberger, C.R. Simmons, N.E. Fahmi, R. Freeman, N. Stephanopoulos*. J. Am. Chem. Soc. (2020) 3:1406-1416. (selected as “ACS Editor’s Choice” article)
Progress toward sourcing plants for new bioconjugation tools: a screening evaluation of a model peptide ligase using a synthetic precursor. T. Mahatmanto*, I. Azizah, A, Buchberger, N. Stephanopoulos. 3 Biotech (2019) 9:442.

Peptide–Oligonucleotide Hybrid Molecules for Bioactive Nanomaterials. N. Stephanopoulos*. Bioconj. Chem. (2019) 30:1915-1922.

Strategies for stabilizing DNA nanostructures to biological conditions. N. Stephanopoulos* . ChemBioChem (2019) 20:2191-2197.

Tunable nanoscale cages from self-assembling DNA and protein building blocks. Y. Xu, S. Jiang, C. Simmons, R.P. Narayanan, F. Zhang, A.-M. Aziz, H. Yan, N. Stephanopoulos*. ACS Nano (2019) 13:3545–3554.

Label-Free Detection of Conformational Changes in Switchable DNA Nanostructures with Microwave Microfluidics. A. Stelson, M. Liu, C.A.E. Little, C.J. Long, N.D. Orloff, N. Stephanopoulos*, J.C. Booth*. Nat. Commun. (2019) 10:1174.

Emerging applications of peptide–oligonucleotide conjugates: bioactive scaffolds, self-assembling systems, and hybrid nanomaterials. T. MacCulloch, A. Buchberger, N. Stephanopoulos*. Org. Biomol. Chem. (2019) 17:1668-1682.

Rapid Photoactuation of a DNA Nanostructure using an Internal Photocaged Trigger Strand. M. Liu, S. Jiang, O. Loza, P. Sulc, N.E. Fahmi, N. Stephanopoulos*. Angew. Chem. Int. Ed. (2018) 57:9341 –9345. (selected as paper for Wiley’s Joint Special Collection on Biopolymers, for the Murray Goodman Award Symposium at the 2019 ACS Spring Meeting: bit.ly/wileybiopolymers19)
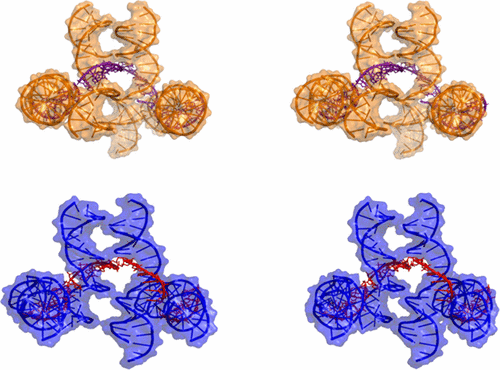
Tuning the Cavity Size and Chirality of Self-Assembling 3D DNA Crystals. C. Simmons, F. Zhang, T. MacCulloch, N.E. Fahmi, N. Stephanopoulos, Y. Liu, N. Seeman, H. Yan*, J. Am. Chem. Soc. (2017) 139:11254-11260.

A Robust Vintronectin-Derived Peptide for the Scalable Long-term Expansion and Neuronal Differentiation of Human Pluripotent Stem Cell (hPSC)-derived Neural Progenitor Cells (hNPCs). D. Varun, G.R. Srinivaan, Y.-H. Tsai, H.-J. Kim, J. Cutts, F. Petty, R. Merkley, N. Stephanopoulos, D. Dolezalova, M. Marsala, D.A. Brafman*. Acta Biomater. (2017) 48:120-130.
Prior to ASU:
(* = co-first authors)

Reversible self-assembly of superstructured networks. R. Freeman, M. Han, Z. Álvarez, J.A. Lewis, J.R. Wester, N. Stephanopoulos, M.T. McClendon, C. Lynsky, J.M. Godbe, H. Sangji, E. Luijten, S.I. Stupp. Science (2018) 362:808-813.

Electrophysiological assessment of a peptide amphiphile nanofiber nerve graft for facial nerve repair. J.J. Greene, M.T. McClendon, N. Stephanopoulos, S.I. Stupp, C.-P. Richter. J. Tissue Eng. Regen. Med. (2018) 12:1389–1401.
Creating a stem cell niche in the inner ear using self-assembling peptide amphiphiles. A.J. Matsuoka , Z.A. Sayed, N. Stephanopoulos, E.J. Berns, A.R. Wadhwani, Z.D. Morrissey, D.M. Chadly, S. Kobayashi, A.N. Edelbrock, T. Mashimo, C.A. Miller, T.L. McGuire, S.I. Stupp, J.A. Kessler. PLoS ONE (2017) 12:e0190150.
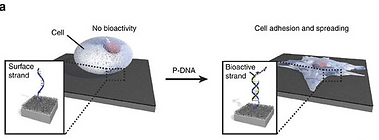
Instructing cells with programmable DNA-peptide hybrids. R. Freeman*, N. Stephanopoulos*, Z. Álvarez, J.A. Lewis, S. Sur, C.M. Serrano, J. Boekhoven, S.S. Lee, S.I. Stupp. Nat. Commun. (2017) 8:15982
The Powerful Functions of Peptide-Based Bioactive Matrices for Regenerative Medicine. C. Rubert-Perez, N. Stephanopoulos, S.S. Lee, S. C. Newcomb, Sur, S.I. Stupp. (invited review) Ann. Biomed. Eng. (2015) 43:501-514.
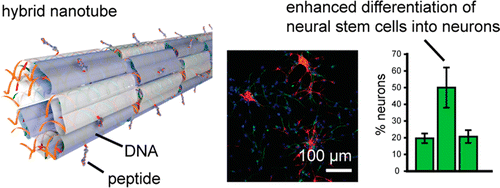
Bioactive DNA-Peptide Nanotubes Enhance the Differentiation of Neural Stem Cells Into Neurons. N. Stephanopoulos, R. Freeman, H.N. Scheler, S. Sur, S. Jeong, F. Tantakitti, J.A. Kessler, S.I. Stupp. Nano Lett. (2015) 15:603-609.

A bioengineered peripheral nerve construct using aligned peptide amphiphile nanofibers. A. Li, A. Hokugo, A. Yalom, E.J. Berns, N. Stephanopoulos, M.T. McClendon, L.A. Segovia, I. Spigelman, S.I. Stupp, R. Jarrahy. Biomaterials (2014) 35:8780-8790.
Self-Assembly for the Synthesis of Functional Biomaterials. N. Stephanopoulos, J.H. Ortony, S.I. Stupp. (invited review) Acta Materialia (2013) 61:912-930.

Antibody-guided photoablation of voltage-gated potassium channels. J. Sack, N. Stephanopoulos, D.C. Austin, M.B. Francis, J.S. Trimmer. J. Gen. Physiol. (2013) 142:315-324.
Choosing an Effective Protein Bioconjugation Strategy. N. Stephanopoulos, M.B. Francis. (invited review) Nat. Chem. Biol. (2011) 7:876-884.
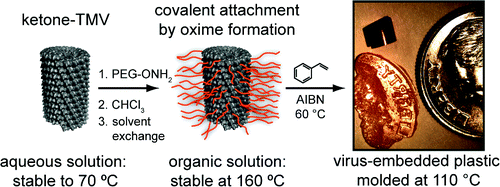
Dramatic Thermal Stability of Virus-Polymer Conjugates in Hydrophobic Solvents. P.G. Holder, D.T. Finley, N. Stephanopoulos, R. Walton, D.S. Clark, M.B. Francis. Langmuir (2010) 26:17383–17388.

Dual-Surface Modified Virus Capsids for Targeted Delivery of Photodynamic Agents to Cancer Cells. N. Stephanopoulos, G.J. Tong, S.C. Hsiao, M.B. Francis. ACS Nano (2010) 4:6014-6020.
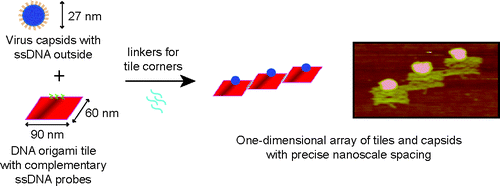
Immobilization and One-Dimensional Arrangement of Virus Capsids with Nanoscale Precision Using DNA Origami. N. Stephanopoulos*, M. Liu*, G.J. Tong, Z. Li, Y. Liu, H. Yan, M.B. Francis. Nano Lett. (2010) 10:2714-2720.
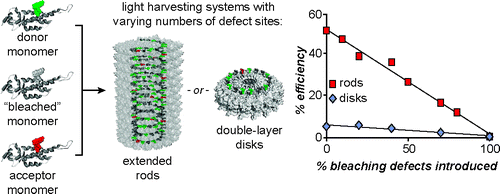
The Impact of Assembly State on the Defect Tolerance of TMV-based Light Harvesting Arrays. R.A. Miller, N. Stephanopoulos, J.M. McFarland, A.S. Rosko, P.L. Geissler, M.B. Francis. J. Am. Chem. Soc. (2010) 132:6068-6074.
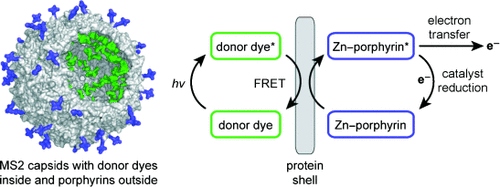
Nanoscale Integration of Sensitizing Chromophores and Porphyrins Using Bacteriophage MS2. N. Stephanopoulos, Z.M. Carrico, M.B. Francis. Angew. Chem. Int. Ed. (2009) 121:9662-9666.
Nanoscale process systems engineering: Toward molecular factories, synthetic cells, and adaptive devices. N. Stephanopoulos, E.O.P. Solis, G. Stephanopoulos. (invited perspective) AIChE J. (2005) 51:1858-1869.
Invited book chapters
(*= corresponding author)
N. Stephanopoulos*, R. Freeman*, “DNA-based materials as self-assembling scaffolds for interfacing with cells”, “Self-Assembling Biomaterials: Molecular Design, Characterization and Application in Biology and Medicine, 1st Edition” 2018, pp. 157-175. (Elsevier)
L. Avolio, D. Sipes, N. Stephanopoulos, S. Sur*, “Recreating stem-cell niches using self-assembling biomaterials”, “Self-Assembling Biomaterials: Molecular Design, Characterization and Application in Biology and Medicine, 1st Edition” 2018, pp. 421-454. (Elsevier)
N. Stephanopoulos, M.B. Francis*, “Making New Materials from Viral Capsids”, “Polymer Science: A Comprehensive Reference, 1st Edition” 2012, Vol. 9, pp. 247-266. (Elsevier)
